Overview
Ready-to-use reaction mix designed specifically for applying the SYBR Green I detection format in the LightCycler® 480 Multiwell Plates, or LightCycler® 8-Tube Strips on the LightCycler® PRO or LightCycler® 96 Instrument. It is used to perform hot start PCR. Hot start PCR has been shown to significantly improve the specificity and sensitivity of PCR by minimizing the formation of nonspecific amplification products at the beginning of the reaction. FastStart Taq DNA Polymerase is a chemically modified form of thermostable recombinant Taq DNA polymerase that shows no activity up to +75°C. The enzyme is active only at high temperatures, where primers no longer bind nonspecifically. The enzyme is completely activated (by removal of blocking groups) in a single pre-incubation step (+95°C, 5 to 10 minutes) before cycling begins. Activation does not require the extra handling steps typical of other hot start techniques.
Highlights
- Improve specificity, sensitivity, and PCR yield with hot start protocols.
- Achieve consistent, high-quality performance with the LightCycler® 96 and Nano Instruments using Multiwell Plates, 96 or 8-Tube Strips.
- Save time with a convenient, ready-to-use 2x concentrated hot start master mix
How this product works
The basic steps of DNA detection by SYBR Green I during real-time PCR on the LightCycler® Systems are:
- At the beginning of amplification, the reaction mixture contains the denatured DNA, the primers, and the dye. The unbound dye molecules weakly fluoresce, producing a minimal background fluorescence signal, which is subtracted during computer analysis.
- After annealing of the primers, a few dye molecules can bind to the double strand. DNA binding results in a dramatic increase of the SYBR Green I molecules’ light emission upon excitation.
- During elongation, more and more dye molecules bind to the newly synthesized DNA. If the reaction is monitored continuously, an increase in fluorescence is viewed in real time. Upon denaturation of the DNA for the next heating cycle, the dye molecules are released and the fluorescence signal falls.
- Fluorescence measurement at the end of the elongation step of every PCR cycle is performed to monitor the increasing amount of amplified DNA.
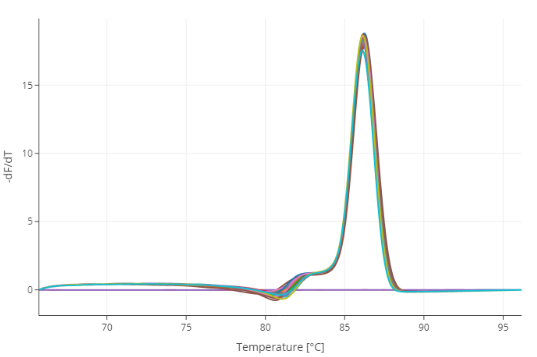
Figure: Melting curve analysis of amplified samples with cDNA dilutions of 1,200 copies, 4,900 copies, 20,000 copies, 78,000 copies, and 310,000 copies per well.








