The Seven steps of sample prep.
Converting a sample to a sequencing library typically involves seven steps, and every step of the process has the potential to reduce or bias the value. To preserve sample integrity and convert nucleic acids into sequenceable molecules with minimal loss, you have to work with the best sample prep solutions for each step.
Next-generation sequencing
The seven steps of your workflow.
Roche Sample Prep Solutions encompass all the steps required to convert a sample to a sequencing-ready library. From sample collection to library quantification, we offer high-quality products and integrated workflow solutions for different sample types and sequencing applications that are proven, complete and simple.
Step 1: Sample Collection
Sample collection is a critical preanalytical step for any application. Investigations on the influence of preanalytical considerations such as sample collection and storage have underscored their importance in determining the validity and usefulness of data for downstream analyses. In order to obtain the most clinically relevant information, it is imperative to ensure that there is minimal sample loss, consistency in collection, and preservation of sample integrity.
The Cell-Free DNA Collection Tube* is a direct-draw tube for the collection, stabilization, and transportation of whole blood specimens. It effectively prevents blood coagulation and cell lysis, and it preserves nucleated cells to enable efficient analysis of cell-free DNA (cfDNA). This product is available only as part of our NIPT and liquid biopsy workflows.
* Available only for use with Roche products in the context of liquid biopsy and NIPT
Step 2: Sample Enrichment
The AVENIO Millisect System is an automated, high-performance tissue dissection system that enables precise and consistent recovery of formalin-fixed paraffin-embedded (FFPE) tissue for molecular pathology. The system is optimized to fit easily into a variety of clinical workflow configurations. It improves diagnostic testing results by reducing false negatives and can help you efficiently extract more clinically relevant information from every sample.
Step 3: Sample Preparation (Nucleic acid extraction)
In genomics, nucleic acid extraction is the starting point to unlock the intrinsic value in every sample. Sample preparation is a critical step for next-generation sequencing (NGS): samples are precious and preparing NGS-compatible genomic DNA or RNA with high yield and high quality is crucial for the success of many clinical research and diagnostic applications.
The Roche family of MagNA Pure Systems automates and simplifies nucleic acid extraction, which dramatically reduces the handling errors associated with manual techniques. The MagNA Pure Systems have been optimized for NGS workflows using new protocols compatible with Roche Sample Prep Solutions, bringing you confidence in your sequencing results.
In the new KAPA HyperPETE (Primer Extension Target Enrichment) workflow, a separate DNA Extraction kit is provided for the extraction of PCR-ready DNA from FFPE samples. The KAPA NGS FFPE DNA Polishing kit reduces sequencing errors originating from deaminated cytosines – a common challenge in FFPE-treated samples. For the final quality control, the KAPA NGS FFPE QC kit can be used.
Step 4: Sample Quantification/QC
A major challenge in Next-Generation Sequencing (NGS) is the ability to process low-input samples of variable quality with predictable success rates. Because these samples are typically very precious, the best sample prep solutions are needed to convert the available DNA into sequenceable molecules with minimal loss and bias.
Being able to assess the probability of sequencing success from challenging samples prior to library construction not only reduces sample processing time and cost but also helps set expectations related to data quality. Commonly used sample QC methods based on spectrophotometry or fluorometry are typically insufficient to predict the yield and quality of library yields prepared from degraded DNA samples.
The KAPA hgDNA Quantification and QC Kits offer a complete, qPCR-based kit for the quantification and quality assessment of human genomic DNA samples in a single assay.
Step 5: Library Preparation & Amplification
Successful sequencing requires high-quality library construction of sufficient yield, where the workflow depends on the NGS application. This includes a series of complex steps: Input sample QC, DNA or RNA library construction and target enrichment (optional), library amplification, and library quantification prior to sequencing.
Because of the number of steps involved and their inherent complexities, each step can potentially introduce errors or biases that could significantly impact the success and reliability of sequencing results. Therefore, the use of high-quality reagents and optimized protocols are required to obtain superior sequencing libraries with the lowest bias, and the highest and most uniform coverage.
Roche offers a range of sample preparation reagents and high-quality enzymes engineered through our Directed Evolution technology for every step of the NGS sample preparation workflow.
DNA: Library Preparation
DNA library preparation for Next-Generation Sequencing (NGS) involves fragmentation of target DNA molecules into varying sizes, end-repair & A-tailing, and ligation of platform-specific adapters to the library. The libraries are then amplified and cleaned up subsequently. Depending on the nature of the DNA sample (e.g. fresh sample versus formalin-fixed paraffin embedded (FFPE) tissue), the workflow may require optimization.
Fragmentation of DNA can be achieved through mechanical or enzymatic shearing. While mechanical shearing is considered the gold standard, enzyme-based fragmentation affords speed, less hands-on time, and convenience to enable automation of the workflow. Since the adapter size is constant, the library size is adjusted based on the desired insert size and the type of application.
The KAPA Library Preparation Kits include modules required for end-repair, A-tailing, ligation, and amplification. KAPA HiFi DNA polymerase has been optimized to reduce bias during amplification to ensure uniformity of coverage across the genome. With KAPA Adapters and Cleanup Beads, Roche offers a complete library preparation solution for NGS.
Benefits of the KAPA Library Preparation Kits
- Compatibility with a wide variety of DNA inputs and sample types offering high yields of adapter-ligated library molecules
- Reduced PCR amplification bias resulting in improved sequencing coverage
- Flexible kit formats to allow for PCR-free workflows
- High-quality library construction reagents supplied in convenient master-mix formats
- Kits are compatible with the Illumina sequencing platform and provide qualified automated methods.
KAPA HyperPrep Kits offer a streamlined mechanical shearing library preparation protocol, combining several enzymatic steps and eliminating bead cleanups to reduce library preparation time and improve consistency. The novel, single-tube chemistry offers further improvements to library construction efficiency, particularly for challenging samples such as FFPE and cell-free DNA. KAPA HyperPrep Kits offer complete library preparation solutions with KAPA Adapters* and KAPA Pure Beads*.
KAPA HyperPlus Kits, our most advanced library preparation option, provide a streamlined workflow that includes fully automatable fragmentation and library preparation in a single tube. This integrated solution combines enzymatic fragmentation, with the quality of mechanical shearing, and the speed and convenience of tagmentation-based workflows.
KAPA EvoPlus is our newest kit on the block and will make you rethink your perception of enzymatic fragmentation. It combines the speed and efficiency of enzymatic fragmentation with the exceptional performance and robustness of mechanical shearing, taking the best of both worlds to form the new gold standard in library preparation.
With a simplified workflow, automation-friendly plated ready mixes, and less time-consuming prep steps, labs can now focus their resources elsewhere whilst having the peace of mind about the quality of results. All that comes with the usual outstanding single vendor support across the entire workflow. KAPA HyperPrep Kits offer complete library preparation solutions with KAPA Adapters* and KAPA (Hyper)Pure Beads*.
* These marked products are also available as stand-alone products.
RNA: Library Preparation
RNA sequencing (RNA-Seq) leverages Next-Generation Sequencing (NGS) to provide insights into global transcriptional events, coding mRNA transcripts or the profiles of RNA species, such as lncRNA. RNA library preparation for NGS utilizes specific strategies, such as enrichment of mRNA or depletion of rRNA, based on the objective of the sequencing experiment.
Roche Sequencing Solutions offer workflows for different sample types and RNA-Seq applications that are proven, simple and complete. This portfolio of KAPA RNA HyperPrep Kits contain high-quality enzymes, including KAPA HiFi DNA polymerase, developed through our Directed Evolution Technology for constructing RNA libraries with minimal GC bias and uniform sequence coverage.
The KAPA RNA HyperPrep Kits utilize a novel chemistry to enable the combination of enzymatic steps and fewer reaction purifications. This results in a truly streamlined solution for the preparation of high-quality RNA-seq libraries. The strand-specific workflow is flexible, supporting library construction from lower-input amounts and degraded samples. It is compatible with mRNA capture as well as ribosomal depletion.
Benefits of KAPA RNA HyperPrep Kits
- Single-day library construction for Illumina sequencing, inclusive of RNA enrichment
- Robust performance across different sample types and input amounts
- Higher success rates with low-input and degraded samples
- Integrated service and support for the entire workflow from RNA to sequencing-ready library
- Validated KAPA RNA HyperPrep Kit data analysis solutions provided through the Genialis platform
KAPA RNA HyperPrep Kits: Specifications

Library amplification
Most next-generation sequencing (NGS) library construction workflows include one or more PCR amplification steps to add functional elements (e.g., sample indices, molecular barcodes or flow cell oligo binding sites), enrich for sequencing-competent DNA fragments and/or generate a sufficient amount of library material for downstream processing. PCR is also applied for other purposes in NGS workflows. Whenever PCR is performed, care must be taken to select appropriate enzyme and protocol parameters, to ensure high efficiency, minimize amplification bias and limit the introduction of sequencing artifacts.
Roche Sequencing Solutions offers a family of KAPA HiFi Kits containing KAPA HiFi DNA Polymerase, engineered using Directed Evolution technology. The KAPA HiFi enzyme contributes to higher overall sequencing quality and economy by offering high efficiency, low bias amplification of DNA and cDNA for both NGS and PCR applications. This translates to more uniform coverage of difficult regions and lower duplication rates allowing researchers to unlock the potential of their samples. In addition, the KAPA HiFi family includes KAPA HiFi Uracil+ Kits, which contain a uracil-tolerant variant, KAPA HiFi Uracil+ DNA Polymerase. This enzyme still offers the same benefits enabled by KAPA HiFi, but is also tolerant to amplification of modified nucleic acids, including bisulfite-converted DNA.
Benefits of KAPA HiFi Kits
- Higher and more uniform coverage with lower drop out of difficult regions
- Higher yields, low duplication rates and fewer wasted sequencing reads
- Higher success rates with different sample types/applications
- Convenience with the consistency of one core enzyme
- Trusted as shown through over 10 years in the industry as well as thousands of peer-reviewed publications
KAPA Library Amplification Kits include KAPA HiFi DNA Polymerase, a novel enzyme engineered using our directed evolution technology for ultra-high fidelity and robustness. KAPA HiFi has become the enzyme of choice for NGS library amplification due to its ability to amplify complex DNA populations with high fidelity, high efficiency and very low amplification bias. This results in lower duplication rates and improved coverage of GC- and AT-rich regions, promoters, low-complexity and other challenging regions.
KAPA HiFi Uracil+ DNA Polymerase is a modified version of KAPA HiFi DNA Polymerase. It is engineered to tolerate uracil residues in bisulfite-treated DNA. Bisulfite treatment of DNA results in the conversion of unmethylated cytosine residues into uracil, while the methylated residues are left unmodified. KAPA HiFi Uracil+ DNA Polymerase has been developed to read through uracil residues while still retaining the performance benefits of HiFi DNA Polymerase, such as high yields, low-bias, and uniform sequencing coverage.
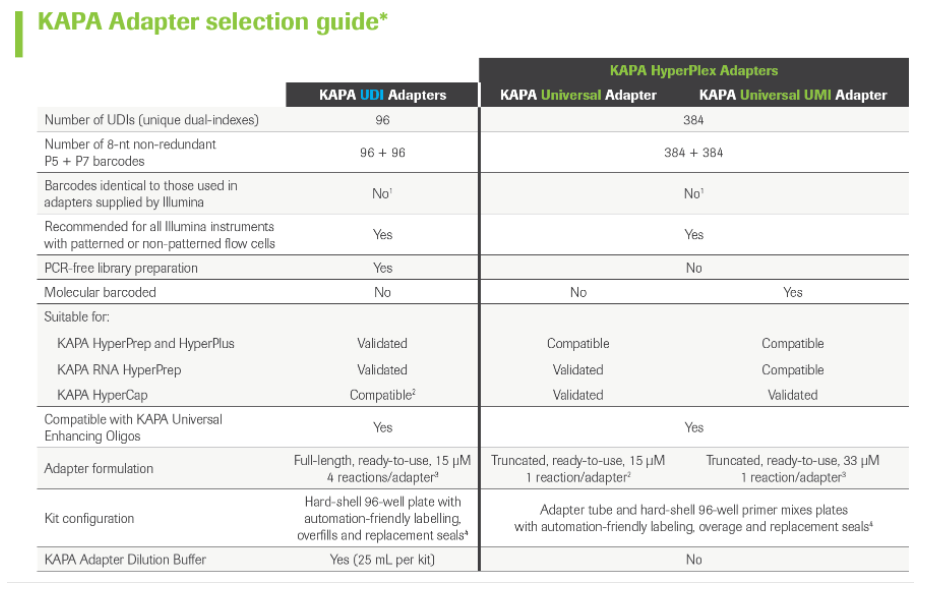
KAPA Unique Dual-Indexed Adapters are high-quality, ready-to-use, full-length adapters for ligation-based library construction in Illumina® sequencing workflows. KAPA Unique Dual-Indexed (UDI) Adapters comprise 96 adapters with non-redundant index combinations to mitigate the impact of index misalignment. Roche Universal Blocking Oligo Kits eliminate the need for adapter-matched blocking oligos, affording a simple and faster workflow.
As part of the new and validated KAPA HyperCap (v3) workflow, the KAPA Universal Adapter is a short adapter with T-overhangs that ligates to library fragments coming out of the A-tailing step of the workflow. The standard version carries no indexes, while the KAPA Universal UMl Adapter includes unique molecular identifiers that improve correct duplicate filtering and thereby increase sensitivity, especially in low-input applications like somatic oncology.
When using KAPA Universal Adapters, sample barcodes have to be incorporated in the following library amplification step by using KAPA UDI Primer Mixes. These primer mixes include forward and reverse primers and carry unique 8-nucleotide indices that allow for library pooling before target capture and sequencing.
KAPA Universal Adapters together with KAPA UDI Primer Mixes replace KAPA UDI Adapters in version 3 of the HyperCap workflow.
KAPA Adapters selection guide:
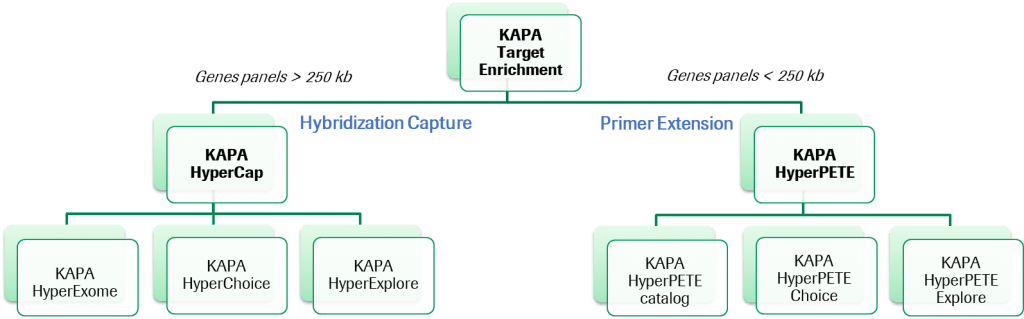
Step 6: Target Enrichment
Target Enrichment is a cost-effective and efficient method to capture specific regions of interest after library preparation for NGS and has many advantages over Whole-Genome Sequencing (WGS):
Target enrichment enables focused sequencing resources, which leads to reduced cost and simplified analysis. The technique increases the probability of sequencing DNA fragments of the targeted regions compared to all other genes in the genome. As a result, the rest of the genome can be disregarded, simplifying downstream bioinformatics analysis. It also provides greater sequencing depth, which helps prevent false interpretations of sequencing data. Furthermore, it increases the ability of NGS to detect variants from limited sample amounts and increases sensitivity. Because of this sequencing depth and sensitivity, target enrichment is valuable in variant calling in cancer research, identifying disease-associated mutations, single nucleotide variants (SNPs), single-gene disorders and in gene expression studies.
Several strategies are employed for target enrichment and enriching specific genomic regions of interest before sequencing. The most commonly used methods are hybrid capture enrichment and amplicon-based enrichment.
Roche Sequencing Solutions offers hybridization-based target enrichment solutions that are a part of an integrated HyperCap workflow, which incorporates both high-quality probes as well as the streamlined KAPA Library Prep Kits. This workflow enables focusing sequencing resources for targeted resequencing applications in human genetic disease and cancer research.
Hybridization-Based Target Enrichment
Targeted sequencing enables researchers to enrich specific genes, exons and/or other genomic regions to allow sequencing reads to be dedicated to only the regions that are of interest, which results in time and cost savings. To enrich specific target regions for Next-Generation Sequencing (NGS) and bioinformatic analysis, hybridization or capture-based target enrichment is a frequently used method. This technique can be applied to single nucleotide polymorphism (SNP) detection, insertion/deletion (indel) detection, copy number variation (CNV) detection, and structural variation detection.
KAPA Target Enrichment Portfolio
Now available is Roche’s new KAPA Target Enrichment Portfolio — KAPA HyperExome, KAPA HyperChoice and KAPA HyperExplore probes. Our industry-leading selection algorithm of probes will now offer higher performance, higher probe fidelity and in-solution hybridization capture, as well as longer probes with fixed length.
Benefits of KAPA Target Enrichment Probes
- Better capture uniformity
- Lower duplicate rate
- Higher target coverage
- Custom design expertise
- KAPA HyperExome ProbesThe KAPA HyperExome Probes are Roche’s brand new Whole Exome Sequencing solution, based on the next generation KAPA Target Enrichment Probe’s technology. Their design and technology provide broad, relevant database coverage and high sequencing efficiency.The product has been extensively optimized through the design and manufacturing process to increase coverage in hard-to-sequence regions for a more uniform and complete exome. Based on KOL feedback, Roche Sequencing scientists created the KAPA HyperExome Probes, a sequencing efficient design with actionable and well-annotated content.KAPA HyperExome Probes are targeting the GRCh38/hg38 genome assembly (coordinates for hg19 annotation are also provided) by covering the CCDS, RefSeq, Ensembl, GENCODE and ClinVar genomic databases in an efficient ~43 Mb capture target size, including 387 sample tracking SNPs to streamline sample identity tracking throughout the WES workflow. Designed for greater sequencing depth and higher uniformity with low PCR duplicates.Features and benefits of KAPA HyperExome Probes:
-
-
- Better by design– Uncover difficult regions and access more content from important genomic databases
- Validated with the KAPA HyperCap Workflow v3.0 for high sequencing efficiency by superior uniformity and low duplication rates
- Higher result confidencewith >98.7% sensitivity and >99.7% specificity of SNP detection
- Stronger enrichment by high fidelity probes that are manufactured with KAPA HiFi DNA Polymerase and target both DNA strands
- Streamline your sample identity tracking to new levels of result confidence by intrinsic targeting 387 sample tracking SNPs
- Experience consistent quality and efficiency with NGS probe pool QC and larger reaction pack sizes
- Experience the convenience of fully integrated, validated and supported workflow from one provider
-
- KAPA HyperCap Fixed Panels Based on our HyperCap design and probe technology, we’re offering two high-performing off-the-shelf panels with relevant content and reduced delivery times (compared to custom panels).The KAPA HyperCap Heredity panel is a 10 Mb capture target panel covering the full CDS of 3332 genes strongly associated with inherited disease, accounting for the majority of the total ClinVar documented pathogenic and likely pathogenic variant content. It provides an optimal balance of relevance and sequencing efficiency.The KAPA HyperCap Oncology panel is a 214 Kbp capture target panel covering the full CDS of 13 genes related to somatic oncology research (ATM, BRCA1, BRCA2, BRIP1, CHEK2, EGFR, ERBB2, KRAS, MET, MYCN, RAD51C, RAD51D, TP53) and hotspot variants across 67 genes.
- KAPA Target Enrichment Custom Probes Roche introduces its target enrichment custom probes, KAPA HyperChoice and KAPA HyperExplore, for improved target capture NGS. KAPA HyperChoice Probes enable custom human probe design up to 200 Mb, while HyperExplore Probes are for non-human or more complex custom designs pushed beyond normal boundaries. HyperDesign, a new online tool that uses Roche’s probe design algorithm and selection, allows you to customize target enrichment panels to meet the unique needs of your research.Features and Benefits of KAPA Target Enrichment Probes (KAPA HyperChoice and KAPA HyperExplore Probes):
- Better by design, using Roche’s renowned probe design and selection algorithms, enable access to challenging genomic regions.
- Seamless online generated designs in HyperDesign either with a few clicks or through an expert designer-assisted process
- Stronger enrichment with high fidelity probes manufactured using KAPA HiFi DNA Polymerase
- Consistent quality and performance with probe presence and concentration QC by NGS
- High uniformity and low duplication for better sequencing efficiency with less optimization
Primer Extension Target Enrichment
Launched in 2021, KAPA HyperPETE stands for Primer Extension Target Enrichment which represents a new target enrichment technology. KAPA HyperPETE combines the workflow benefits of amplicon-based enrichment with the performance of probe capture approaches. KAPA HyperPETE:
- Its technology enables high performing capture in a single day
- Has a simple, easy-to-use, and automation-friendly workflow
- Performs better than amplicon technology
KAPA HyperPETE is a small-panel specialist, ideal for somatic oncology research, RNA Fusion, and germline panels under 250KB.
A set of predefined panels (Pan Cancer, Hot Spot, Lung Cancer Fusion, Hereditary Oncology , Newborn Screening) are complemented by a custom panel option for both DNA and RNA targets in humans (HyperPETE Choice) and other organisms (HyperPETE Explore). The design of these custom panels is possible through our existing HyperDesign platform.
To drive more efficient and accurate variant identification, KAPA HyperPETE Panels are supported by an integrated secondary analysis solution, NAVIFY Mutation Caller.
Step 7: Library Quantification/QC
Using a qPCR-based solution, KAPA Library Quantification Kits provide accurate and reliable quantification of libraries prepared for sequencing on Illumina and IonTorrent platforms, across a wide range of library types, concentrations, fragment distributions and GC content. This step does not only represent a final QC of the libraries but also ensures that they can be pooled according to their concentrations on the sequencer.
Kits include KAPA SYBR FAST qPCR Master Mix, a platform-specific library quantification primer premix, and a pre-diluted set of DNA standards.
KAPA and SeqCap are for Research Use Only. Not for use in diagnostic procedures. Millisect and MAGNA PURE 24 and 96 products are for use for in vitro diagnostics Cell-Free DNA Collection Tubes are available both in RUO* and CE-IVD** formats.
*For Research Use Only. Not for use in diagnostic procedures.
**Available for countries that accept CE-Mark.]
Talk to a local expert
We offer proven, simple and complete solutions to optimise your sequencing resources. How can this help you enhance sample prep? We would love to discuss your options. Contact your Roche representative today, and we’ll be in touch.
Roche Sample Prep Solutions
From sample collection to library quantification, our integrated workflow solutions for different sample types and sequencing applications are proven, complete and simple.
Proven
Our portfolio of high-quality sample prep products have served the life sciences, translational and clinical research community for over a decade.
Simple
Streamlining every step of the process, from pipetting to purchasing, improves sequencing success and frees up your time and resources.
Complete
Qualified, complete workflow solutions for a wide range of sample types and applications give you greater peace of mind.